Note about crash last time!
GooFit and LmFit both seem to have compiled code that expects OpenMP - and at least on my Mac, they were linked with different OpenMP libraries (Anaconda's Intel libomp for LmFit, and HomeBrew's libomp for GooFit). So that's why it crashed. I've reinstalled GooFit without OpenMP for now.
GooFit
If you really want to try GooFit, you can try this on OSC:
!pip install --user scikit-build cmake
!PATH=$HOME/.local/bin:$PATH pip install --user --verbose goofitThe extra requirements here are partially to ensure it gets the highest level of optimization, and partially requirements that will eventually go away.
If you are on macOS, scikit-build is broken, you'll need !pip install scikit-build==0.6.1.
from scipy.stats import norm, multivariate_normal
import matplotlib.pyplot as plt
import numpy as np
state = np.random.RandomState(42)
gauss_part = state.normal(1, 2, size=100_000)
lin_part = state.uniform(low=-10, high=10, size=50_000)
total_rand = np.concatenate([gauss_part, lin_part])
import goofit
We need an observable, with a range from -10 to 10.
x = goofit.Observable("x", -10, 10)
We can make an unbinned (or binned) dataset; we need to list the variables it will contain.
data = goofit.UnbinnedDataSet(x)
Let's read in data. GooFit will default to throwing an error if you input a value that is outside the range (-10 to 10 in this case), but we can pass filter=True to simply ignore those values instead.
data.from_matrix([total_rand], filter=True)
If you display a PDF in the notebook, you get a nice pretty-printed version of it's documentation:
goofit.PolynomialPdf
The fitting variables and PDFs need to be setup next.
a = goofit.Variable("a", 0, 0, 1)
linear = goofit.PolynomialPdf("linear", x, [a])
mu = goofit.Variable("mu", 0, -10, 10)
sigma = goofit.Variable("sigma", 1, 0, 5)
gauss = goofit.GaussianPdf("gauss", x, mu, sigma)
We can add multiple PDFs with fractions for each
frac = goofit.Variable("frac", 0.5, 0, 1)
total = goofit.AddPdf("tot", [frac], [gauss, linear])
And, we can fit a PDF to data:
minimum = total.fitTo(data)
The variables are changed in place (mutated):
print(frac)
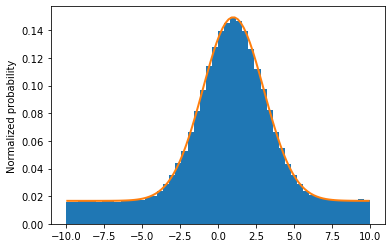
Let's look at a plot.
grid, pts = total.evaluatePdf(x)
xvals = grid.to_numpy().flatten()
fig, ax = plt.subplots(figsize=(6, 4))
ax.hist(total_rand, bins=50, range=(-10, 10), density=True)
ax.plot(xvals, pts, linewidth=2)
# ax.set_xlabel('xvar')
ax.set_ylabel("Normalized probability")
ax.set_ylim(ymin=0)
plt.show()