Review
Week 13 Day 1: Review
| Week | Monday | Wednesday | Friday |
|---|---|---|---|
| 1 | 8-27 Introduction | 8-29 Using Python part 1 | 8-31 Using Python part 2 |
| 2 | 9-03 Labor day | 9-05 OO programming (4) | 9-07 Error accumulation (2) |
| 3 | 9-10 Numerical tools | 9-12 Plotting (3) | 9-14 Using git |
| 4 | 9-17 Random numbers (5) | 9-19 Monte Carlo (5) | 9-21 Project selection |
| 5 | 9-24 Integration rules (6) | 9-26 MC Integration (6) | 9-28 Numerical differentiation (7) |
| 6 | 10-01 Vectorization (8) | 10-03 Linear algebra (8) | 10-05 Linear regression (8) |
| 7 | 10-08 Structured tabular data | 10-10 Cuts and histograms | 10-12 Fall reading days |
| 8 | 10-15 Generating distributions | 10-17 Minimization and fitting | 10-19 Fitting tools |
| 9 | 10-22 Confidence intervals | 10-24 Markov Chain Monte Carlo | 10-26 Performance computing (14) |
| 10 | 10-29 Intro to ODEs (9) | 10-31 Runge–Kutta algorithm (9) | 11-02 Solving ODE problems (9) |
| 11 | 11-05 Fourier Series (10) | 11-07 Fast Fourier Transform (10) | 11-09 Project progress report |
| 12 | 11-12 Veterans day | 11-14 Project progress report | 11-16 Filtering signals (10) |
| 13 | 11-19 Review | 11-21 Student requested topics | 11-23 Thanksgiving |
| 14 | 11-26 Static computation graphs | 11-28 Applied ML topics | 11-30 Sharing and documenting code |
| 15 | 12-03 Term project presentations | 12-05 Term project presentations | 12-07 Term project presentations |
import math
import numpy as np
import numba
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import integrate, optimize, stats, misc
np.random.seed(42)
vals = np.random.normal(0, 1, 1_000_000)
fsummed = math.fsum(vals)
4.1: Integration rules
Integrate the following function. You should use tools provided; don't write your own integrator unless it is necessary for some reason!
$$ f(x) = \int _0 ^1 \sin(\cos x) \, dx \approx 0.73864... $$(From Wolfram Alpha+from+x%3D0+to+1&lk=3))
def f(x):
return np.sin(np.cos(x))
real_deriv = -np.cos(np.cos(0.5)) * np.sin(0.5)
real_deriv
5.3, 8.2, 8.3 : Fitting
We've already done a bit of this, so let's just hit a highlight or two.
Functions:
- Interpolation: connect the dots. Cubic splines were probably the nicest visual interpolation method.
- Linear regression:
np.polyfitornp.linalg.lstsq - Non-linear least squares:
optimize.curve_fit
Distributions:
- Binned fits: See above, be careful and include error
- Unbinned fits: NLL (use
optimize.minimizeor custom package)
Try a quick polyfit here:
np.random.seed(42)
x = np.linspace(0, 2, 200)
y = x * 2 + 0.5 + np.random.normal(0, 0.5, 200)
# Add fit here
plt.plot(x, y, ".")
df = pd.read_csv(
"https://raw.githubusercontent.com/mwaskom/seaborn-data/master/iris.csv"
)
df.head()
Note, we could do this with seaborn:
sns.scatterplot("sepal_length", "sepal_width", "species", data=df)
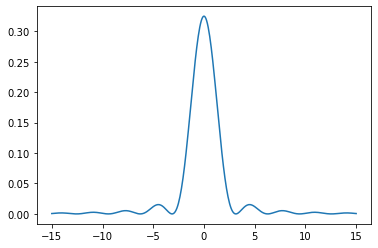
def p(x):
return np.sin(x) ** 2 / x ** 2
Normalization not needed for algorithm, but makes the plots line up:
integ, err = integrate.quad(p, -15, 15.000001)
print(integ, err)
x1 = np.linspace(-15, 15, 400)
y1 = p(x1) / integ
Now compute the metropolis algorithm:
samples = 100_000
sigma = 10
x = np.empty(samples + 1)
x[0] = np.random.rand()
for i in range(samples):
# suggest new position xStar
...
# Compute alpha - the fractional chance of moving to a new point
...
# Accept/reject based on alpha
...
# Add the current (moved?) point
...
plt.plot(x1, y1)
# plt.hist(x[500:], density=True, bins=100)
# plt.yscale('log')
plt.show()
Side project: This is great, but very slow. We can make this a function, and add a couple of @numba.njit's, and get a massive speedup!
We'll start with one, though:
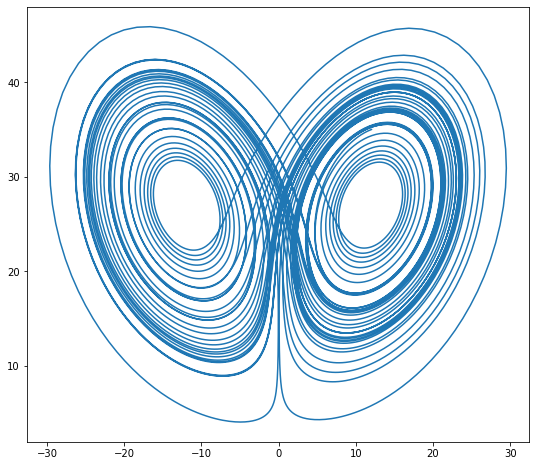
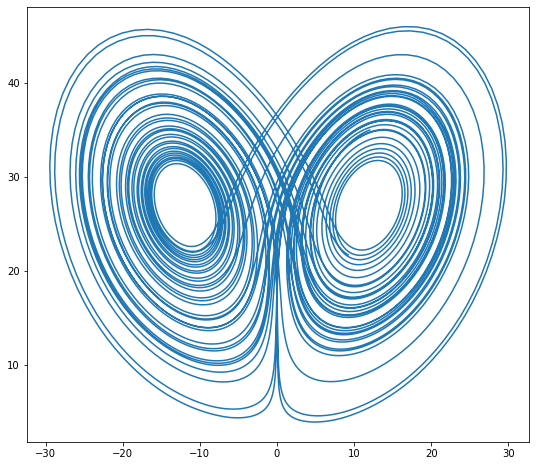
$$ u = \left( \begin{matrix} x \\ y \\ z \end{matrix} \right) $$$$ \dot{u} = \left( \begin{matrix} \sigma (y - x) \\ \rho x - x z - y \\ x y - \beta z \end{matrix} \right) $$Constants given below. (example from Bokeh)
sigma = 10
rho = 28
beta = 8 / 3
theta = 3 * np.pi / 4
def lorenz(t, xyz):
x, y, z = xyz
x_dot = sigma * (y - x)
y_dot = x * rho - x * z - y
z_dot = x * y - beta * z
return np.array([x_dot, y_dot, z_dot])
initial = (-10, -7, 35)
t = np.arange(0, 50, 0.006)
def rk2_ivp(f, init_y, t):
steps = len(t)
order = len(init_y)
y = np.empty((steps, order))
y[0] = init_y
for n in range(steps - 1):
h = t[n + 1] - t[n]
k1 = h * f(t[n], y[n]) # 1.1
k2 = h * f(t[n] + h / 2, y[n] + k1 / 2) # 1.2
y[n + 1] = y[n] + k2 # 1.0
return y
xyz = rk2_ivp(lorenz, initial, t)
xprime = np.cos(theta) * xyz[:, 0] - np.sin(theta) * xyz[:, 1]
plt.figure(figsize=(9, 8))
plt.plot(xprime, xyz[:, 2])
plt.show()
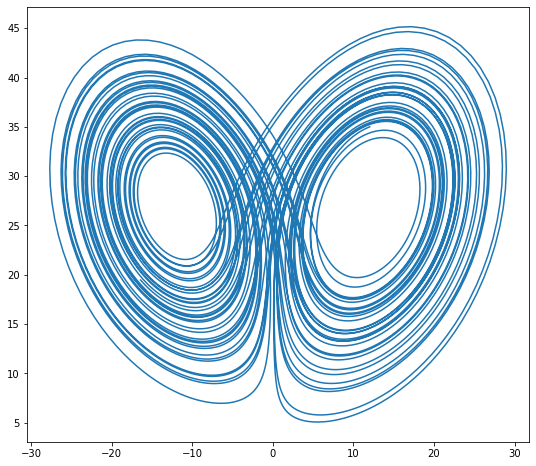
def rk4_ivp(f, init_y, t):
steps = len(t)
order = len(init_y)
y = np.empty((steps, order))
y[0] = init_y
for n in range(steps - 1):
h = t[n + 1] - t[n]
k1 = h * f(t[n], y[n]) # 2.1
k2 = h * f(t[n] + h / 2, y[n] + k1 / 2) # 2.2
k3 = h * f(t[n] + h / 2, y[n] + k2 / 2) # 2.3
k4 = h * f(t[n] + h, y[n] + k3) # 2.4
y[n + 1] = y[n] + 1 / 6 * (k1 + 2 * k2 + 2 * k3 + k4) # 2.0
return y
xyz = rk4_ivp(lorenz, initial, t)
xprime = np.cos(theta) * xyz[:, 0] - np.sin(theta) * xyz[:, 1]
plt.figure(figsize=(9, 8))
plt.plot(xprime, xyz[:, 2])
plt.show()
res = integrate.solve_ivp(lorenz, (0, 100), initial, t_eval=t)
xprime = np.cos(theta) * res.y[0] - np.sin(theta) * res.y[1]
plt.figure(figsize=(9, 8))
plt.plot(xprime, res.y[2])
plt.show()
def lorenz_flip(xyz, t):
return lorenz(t, xyz)
xyz = integrate.odeint(lorenz_flip, initial, t)
xprime = np.cos(theta) * xyz[:, 0] - np.sin(theta) * xyz[:, 1]
plt.figure(figsize=(9, 8))
plt.plot(xprime, xyz[:, 2])
plt.show()