Monte Carlo Integration
import numpy as np
import matplotlib.pyplot as plt
np.zeros((2, 3))
You can also use nested lists or arrays to the normal constructor:
m = np.array([[1, 2], [3, 4]])
print(m)
print(m[0, :], "==", m[0])
m[:, 0]
np.sum(m, axis=1)
np.sum(m, axis=1, keepdims=True)
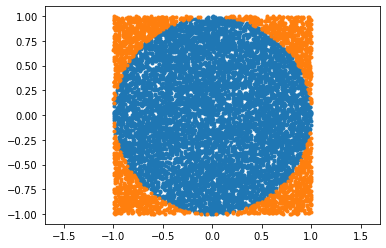
xy = np.random.rand(2, 10000) * 2 - 1
valid = np.sum(xy ** 2, axis=0) < 1
good = xy[:, valid]
bad = xy[:, ~valid]
plt.plot(*good, ".")
plt.plot(*bad, ".")
plt.axis("equal")
np.mean(valid) * 4
xy = np.random.rand(2, 100000) * 2 - 1
r2 = np.sum(xy ** 2, axis=0)
r2[r2 > 1] = 1
print("MC: ", np.mean(np.sqrt(1 - r2)) * 4 * 2)
print("Actual:", 4 / 3 * np.pi)
def f(x):
return np.sum(x, axis=0) ** 2
s = np.random.rand(10, 1000000)
np.mean(f(s))
155 / 6
Error estimate:
155 / 6 * 1 / np.sqrt(s.shape[1])
from scipy import stats
df = 2.74
begin, end = stats.t.cdf([-100, 100], df)
print("Analytic:", end - begin)
x = np.linspace(-100, 100, 1_000)
fig, ax = plt.subplots()
ax.plot(x, stats.t.pdf(x, df))
plt.show()
X = (np.random.rand(1000) - 0.5) * 200
np.mean(stats.t.pdf(X, df)) * 200
1 / np.sqrt(1000)
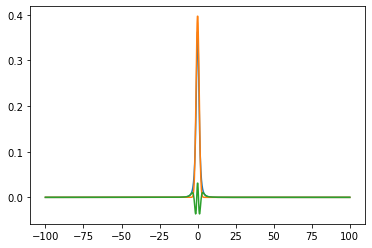
x = np.linspace(-100, 100, 1_000)
fig, ax = plt.subplots()
ax.plot(x, stats.t.pdf(x, df))
ax.plot(x, stats.norm.pdf(x, 0, 1))
ax.plot(x, stats.t.pdf(x, df) - stats.norm.pdf(x, 0, 1.2))
plt.show()
X = (np.random.rand(1000) - 0.5) * 200
Id = np.mean(stats.t.pdf(X, df) - stats.norm.pdf(X, 0, 1.2)) * 200
gaussInt = stats.norm.cdf(100, 0, 1.2) - stats.norm.cdf(-100, 0, 1.2)
print(Id + gaussInt)
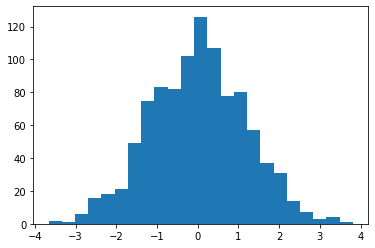
Xg = np.random.normal(0, 1.2, 1000)
Xg = Xg[np.abs(Xg) <= 10]
print(len(Xg))
np.mean(stats.t.pdf(Xg, df) / stats.norm.pdf(Xg, 0, 1.2))
plt.hist(Xg, bins="auto")
plt.show()
Xy = np.random.rand(2, 10000)
Xy[0] -= 0.5
Xy[0] *= 10
plt.plot(*Xy, ".")
plt.xlabel("x")
plt.ylabel("y")
plt.show()
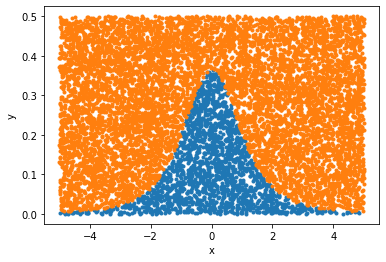
w0 = 0.5 # Must be higher than the maximum of the PDF
Xy[1] *= w0
valid = Xy[1] < stats.t.pdf(Xy[0], df)
plt.plot(*Xy[:, valid], ".")
plt.plot(*Xy[:, ~valid], ".")
plt.xlabel("x")
plt.ylabel("y")
plt.show()
It's not a great way to calculate an integral, but we can:
np.mean(valid) * w0 * 10