Confidence intervals example
Based loosely on A very friendly introduction to confidence intervals
import numpy as np
import scipy.stats as stats
import matplotlib.pyplot as plt
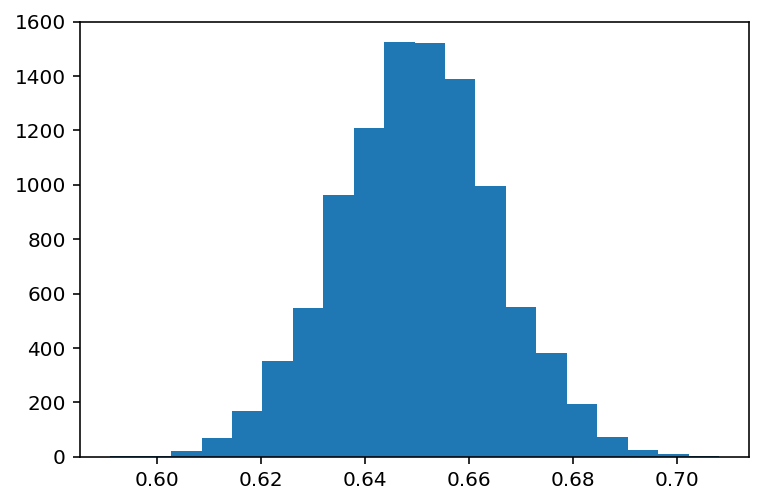
Let's look at taking a small sample of a larger distribution. We'll use the same example given in the reference; this was a poll of the people in the US to see what fraction likes Soccer. Let's assume for the moment we know the real mean, and have the original distribution handy:
love_soccer_prop = 0.65 # Real percentage of people who love soccer
total_population = 325*10**6 # Total population in the U.S. (325M)
# Note: 325e6 would be a floating point number
love_soccer_num = int(love_soccer_prop * total_population)
Bools are smaller than ints, and we can still take the mean, sum, etc. of an array of bools, so we'll use bools. We'll start off with a "sorted" array (people who love Soccer first), we we'll need to be careful when we select samples from it.
population = np.zeros(total_population, dtype=bool)
population[:love_soccer_num] = True
np.mean(population)
for i in range(10):
sample = np.random.choice(population, size=1000)
print(f'Sample {i}: {np.mean(sample)}')
values = np.empty(10_000)
for i in range(len(values)):
values[i] = np.mean(np.random.choice(population, size=1_000))
np.mean(values)
plt.hist(values, bins=20)
plt.show()
confidence=0.95
sem = np.std(sample) / np.sqrt(len(sample)) # scipy.stats.sem(sample)
h = sem * scipy.stats.t.ppf((1 + confidence) / 2, len(sample)-1)
print(f"{np.mean(sample)} +/- {h:.3g} ({confidence:.1%})")
For larger samples (more than 100), we can approximate the student's t distribution with a Gaussian:
sem = scipy.stats.sem(sample)
h = sem * scipy.stats.norm.ppf((1 + confidence) / 2)
print(f"{np.mean(sample)} +/- {h:.3g} ({confidence:.1%})")
In SciPy, we can do this even quicker:
h = scipy.stats.t.interval(confidence, len(sample)-1, loc=np.mean(sample), scale=sem)
print(h)
Anaconda includes another popular package for statistics: StatsModels, which complements scipy.stats and pandas.
import statsmodels.stats.api as sms
inter = sms.DescrStatsW(sample).tconfint_mean()
inter
Physics confidence intervals
Based on the excellent slides here: http://www.phas.ubc.ca/~oser/p509/Lec_16.pdf
For simplicity, we will assume that we are using 90% confidence intervals in the notes below.
In Physics, frequentist confidence intervals are often used for measurements. We make a measurement, then construct a confidence interval from the expected underlying probability distribution for the different possible measurements; we quote the true values for which our measurement is in the 90% range.
This does not tell us that there is a 90% chance the true value is in this range, rather that if we did this experiment many times, 90% of the experiments would contain the true value.
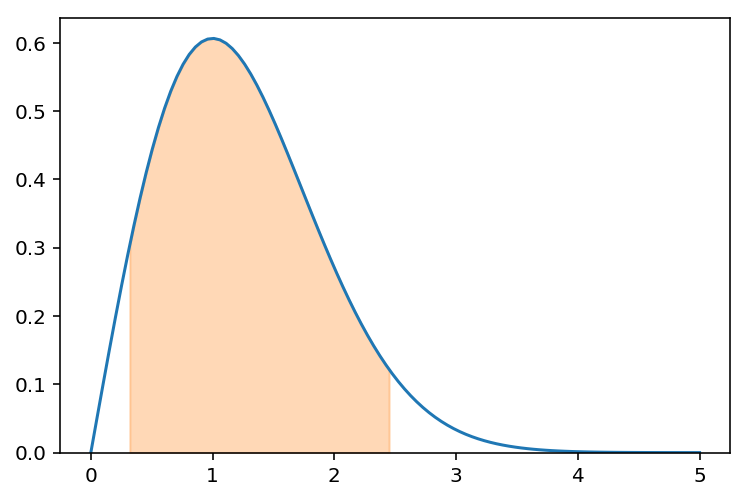
If we have the following distribution, the Neyman confidence interval construction does not specify how to draw the confidence region; any region that contains 90% of the probability could be a 90% confidence interval.
$$ \mathrm{PDF}(x) = x e^{-\frac{1}{2} x^2} $$import sympy as s
s.init_printing()
x = s.symbols('x')
f = x * s.exp(-x**2/2)
f.integrate(x)
This is normalized:
f.integrate((x,0,s.oo))
This is one region:
f.integrate((x,0.32,2.45))
fn = s.lambdify([x], f)
xs = np.linspace(0,5,100)
plt.plot(xs,fn(xs))
xs = np.linspace(0.32,2.45,50)
plt.fill_between(xs, fn(xs), color='C1', alpha=.3)
plt.ylim(ymin=0)
plt.show()
You can also choose an upper limit:
f.integrate((x,0,2.15))
xs = np.linspace(0,5,100)
plt.plot(xs,fn(xs))
xs = np.linspace(0,2.15,50)
plt.fill_between(xs, fn(xs), color='C1', alpha=.3)
plt.ylim(ymin=0)
plt.show()
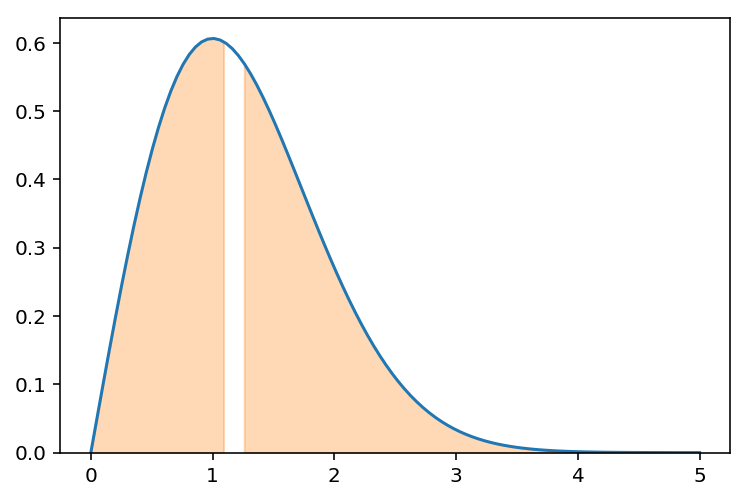
How about this?
f.integrate((x,0,1.09)) + f.integrate((x,1.26,s.oo))
xs = np.linspace(0,5,100)
plt.plot(xs,fn(xs))
xs = np.linspace(0,1.09,50)
plt.fill_between(xs, fn(xs), color='C1', alpha=.3)
xs = np.linspace(1.26,5,50)
plt.fill_between(xs, fn(xs), color='C1', alpha=.3)
plt.ylim(ymin=0)
plt.show()
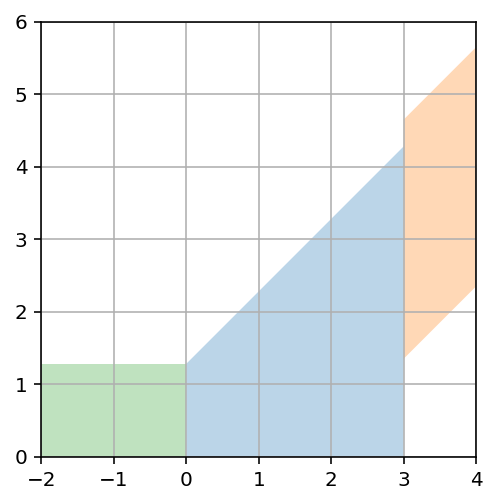
For fixed true value $\mu$, 90% of the probability lies in $\mu-1.28<\hat{\mu}<\infty$. Or $\mu-1.65<\hat{\mu}<\mu+1.65$.
fig, ax = plt.subplots()
# Upper limit quote
x = np.linspace(0,3)
ax.fill_between(x, 1.28 + x, alpha=.3)
# Upper and lower bounds
x = np.linspace(3,4)
ax.fill_between(x, 1.65 + x, x - 1.65, alpha=.3)
# Quote as 0
x = np.linspace(-2,0)
ax.fill_between(x, 1.28 + x*0, alpha=.3)
ax.set_xlim(-2,4)
ax.set_ylim(0,6)
ax.grid()
ax.set_aspect('equal')
plt.show()
From out previous example, $\mu_\mathrm{best}=x$ if $x>0$ or $\mu_\mathrm{best}=0$ if $x\le 0$
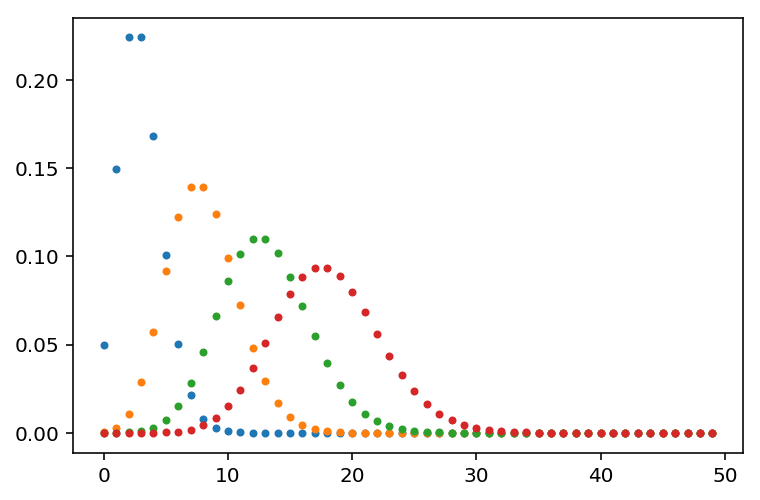
import gammapy.stats as gstats
x_bins = np.arange(0, 50)
mu_bins = np.linspace(0, 15, int(15/0.005) + 1, endpoint=True)
matrix = [stats.poisson(mu + 3.0).pmf(x_bins) for mu in mu_bins]
plt.plot(matrix[0], '.')
plt.plot(matrix[1000], '.')
plt.plot(matrix[2000], '.')
plt.plot(matrix[-1], '.')
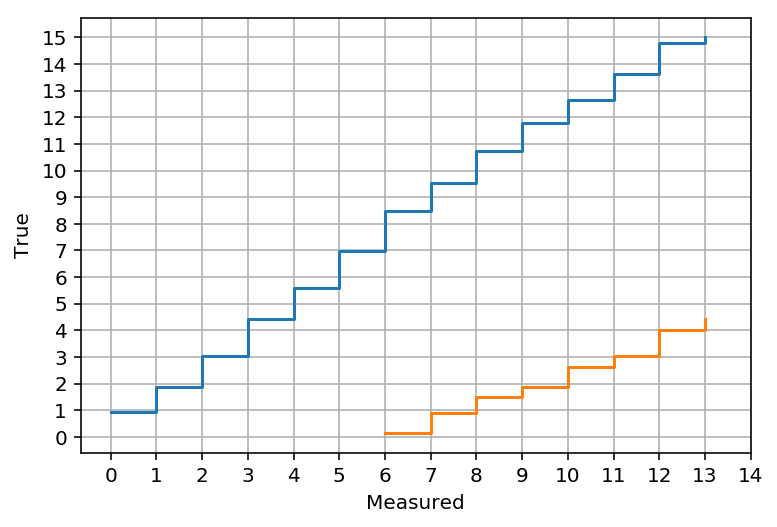
acceptance_intervals = gstats.fc_construct_acceptance_intervals_pdfs(matrix, 0.9)
LowerLimitNum, UpperLimitNum, other = gstats.fc_get_limits(mu_bins, x_bins, acceptance_intervals)
u = np.array([gstats.fc_find_limit(i, UpperLimitNum, mu_bins) for i in range(14)])
l = [gstats.fc_find_limit(i, LowerLimitNum, mu_bins) for i in range(14)]
plt.step(range(14), u, where='post', label='Upper limit')
plt.step(range(14), l, where='post', label='Lower limit')
plt.xticks(range(15))
plt.yticks(range(16))
plt.grid()
plt.xlabel('Measured')
plt.ylabel('True')
plt.show()