Remember to pip install probfit (--user if you are not in a virtual environment). cupy is used at the end; you'll need conda install cupy in an environment. Times are from a 24 core Xeon with a Titan V.
We will put our preperation code in a file, and restart the kernel as we go, just to make sure we don't have any interaction between GPU libraries. likelyhood_answer is defined to be the numpy value - Note that the reduction can produce lots of round of error if not done properly. I know numpy does, not sure about probfit.
Results:
| Package | Hardware | Time per call | Minimize time |
|---|---|---|---|
| Probfit | 24 core Zeon | 1.11 s | 2.23 min |
| Numpy | 24 core Zeon | 21.1 ms | 2.38 s |
| Numba parallel | 24 core Zeon | 11.5 ms | 1.46 s |
| Numba parallel loop | 24 core Zeon | 3.68 ms | 416 ms |
| GooFit | Titan V | 2.30 ms | 333 ms |
| CUPY | Titan V | 2.9 ms | 491 ms |
| CUPY kernel | Titan V | 843 µs | 118 ms |
| CUPY redu | Titan V | 2.50 ms | 343 ms |
| PyTorch | Titan V | 3.68 ms | 594 ms |
| TensorFlow static | Titan V | 1.38 ms | 225 ms |
from read_pretty import read_pretty
read_pretty('common.py')
%run common.py
import matplotlib.pyplot as plt
plt.hist(dist, bins='auto');
import probfit
second_gaussian = probfit.rename(probfit.gaussian, ["x", "mean", "sigma2"])
pdf_function = probfit.AddPdfNorm(probfit.gaussian, second_gaussian)
Now, let's build an unbinned likelyhood function:
unbinned_lh = probfit.UnbinnedLH(pdf_function, dist)
assert unbinned_lh(.5,.5,.5,.5) == likelyhood_answer
%%timeit
unbinned_lh(*np.random.rand(4))
We have to use iMinuit's name based parameter setting interface:
minuit = Minuit(unbinned_lh, **default_params)
And let's do the fit!
%%time
run_and_print(minuit)
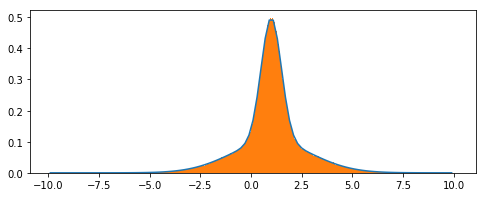
unbinned_lh.draw(minuit);
exit()
%run common.py
import goofit
goofit.GOOFIT_DEVICE
x = goofit.Observable('x', -10, 10)
ds = goofit.UnbinnedDataSet(x)
ds.from_matrix([dist], filter=True)
mean = goofit.Variable('mean', 1.5, -10, 10)
sigma = goofit.Variable('sigma', .4, 0, 1)
sigma2 = goofit.Variable('sigma2', 3, 1, 3)
f_0 = goofit.Variable('f_0', .5, 0, 1)
gauss1 = goofit.GaussianPdf("gauss1", x, mean, sigma)
gauss2 = goofit.GaussianPdf("gauss2", x, mean, sigma2)
pdf = goofit.AddPdf('pdf', [f_0], [gauss1, gauss2])
pdf.fitTo(ds);
Note: On a single thread CPU build, this is about 7-10x faster than probfit. This is on a Titan V GPU, so it's a lot more than that.
Plotting is a bit more verbose, but not too bad:
grid, points = pdf.evaluatePdf(x)
xs = grid.to_matrix().flatten()
fig, ax = plt.subplots(figsize=(8,3))
ax.plot(xs, points)
ax.hist(dist, bins='auto', density=True)
plt.show()
exit()
%run common.py
def gaussian(x, μ, σ):
return 1/np.sqrt(2*np.pi*σ**2) * np.exp(-(x-μ)**2/(2*σ**2))
def add(x, f_0, mean, sigma, sigma2):
return f_0 * gaussian(x, mean, sigma) + (1 - f_0) * gaussian(x, mean, sigma2)
def nll(f_0, mean, sigma, sigma2):
return -np.sum(np.log(add(dist, f_0, mean, sigma, sigma2)))
assert nll(.5,.5,.5,.5) == likelyhood_answer
Note that Numpy uses a good definition of a sum, while Python's built in sum has rounding point errors. This would be more important if we used fewer bits. Let's compare:
# Check reduction sum:
assert -sum(np.log(add(dist, .5, .5, .5, .5))) == likelyhood_answer
# Check floating point accurate sum:
assert -math.fsum(np.log(add(dist, .5, .5, .5, .5))) == likelyhood_answer
%%timeit
nll(*np.random.rand(4))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
# Much (2-3x) slower if we use this in Minuit
# unbinned_lh = probfit.UnbinnedLH(add, dist)
import numba
@numba.njit(parallel=True)
def gaussian(x, μ, σ):
return 1/np.sqrt(2*np.pi*σ**2) * np.exp(-(x-μ)**2/(2*σ**2))
@numba.njit(parallel=True)
def add(x, f_0, mean, sigma, sigma2):
return f_0 * gaussian(x, mean, sigma) + (1 - f_0) * gaussian(x, mean, sigma2)
@numba.njit(parallel=True)
def nll(f_0, mean, sigma, sigma2):
return -np.sum(np.log(add(dist, f_0, mean, sigma, sigma2)))
assert nll(.5,.5,.5,.5) == likelyhood_answer
%%timeit
nll(*np.random.rand(4))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
@numba.jit
def gaussian(x, μ, σ):
return 1/np.sqrt(2*np.pi*σ**2) * np.exp(-(x-μ)**2/(2*σ**2))
@numba.jit
def add(x, f_0, mean, sigma, sigma2):
return f_0 * gaussian(x, mean, sigma) + (1 - f_0) * gaussian(x, mean, sigma2)
@numba.njit(parallel=True)
def nllp(dist, f_0, mean, sigma, sigma2):
s = 0.0
for i in numba.prange(len(dist)):
s += np.log(add(dist[i], f_0, mean, sigma, sigma2))
return -s
def nll(f_0, mean, sigma, sigma2):
return nllp(dist, f_0, mean, sigma, sigma2)
assert nll(.5,.5,.5,.5) == likelyhood_answer
%%timeit
nll(*np.random.rand(4))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
exit()
%run common.py
import cupy
%%time
d_dist = cupy.array(dist)
def gaussian(x, μ, σ):
return 1/cupy.sqrt(2*np.pi*σ**2) * cupy.exp(-(x-μ)**2/(2*σ**2))
def add(x, f_0, mean, sigma, sigma2):
return f_0 * gaussian(x, mean, sigma) + (1 - f_0) * gaussian(x, mean, sigma2)
def nll(f_0, mean, sigma, sigma2):
return -cupy.sum(cupy.log(add(d_dist, f_0, mean, sigma, sigma2)))
assert float(nll(.5,.5,.5,.5)) == likelyhood_answer
%%timeit
float(nll(*np.random.rand(4)))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
Cupy Custom Kernel
Let's go all out and write a CUDA kernel by hand, then call sum on it. We are almost ideal here; the only downside is that we make a single temporary 1M element array before summing.
We could use a reduction kernel, but I don't know if I trust that do not have round off error issues.
mykernel = cupy.ElementwiseKernel(
'float64 x, float64 f_0, float64 mean, float64 sigma, float64 sigma2',
'float64 z', '''
double s12 = 2*sigma*sigma;
double s22 = 2*sigma2*sigma2;
double p = -(x-mean)*(x-mean);
double g = rsqrt(M_PI*s12) * exp(p/s12);
double g2 = rsqrt(M_PI*s22) * exp(p/s22);
z = log(f_0 * g + (1 - f_0) * g2);
''', 'mykernel')
def nll(f_0, mean, sigma, sigma2):
return -cupy.sum(mykernel(d_dist, f_0, mean, sigma, sigma2))
assert float(nll(.5,.5,.5,.5)) == likelyhood_answer
%%timeit
float(nll(*np.random.rand(4)))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
rku = cupy.ReductionKernel(
'float64 x, float64 f_0, float64 mean, float64 sigma, float64 sigma2',
'float64 r',
'''
log( f_0 * rsqrt(2*M_PI*sigma*sigma) * exp(-(x-mean)*(x-mean)/(2*sigma*sigma)) +
(1 - f_0) * rsqrt(2*M_PI*sigma2*sigma2) * exp(-(x-mean)*(x-mean)/(2*sigma2*sigma2)))
''',
'a + b',
'r = -a',
'0',
'redu_kernel'
)
def nll(f_0, mean, sigma, sigma2):
return rku(d_dist, f_0, mean, sigma, sigma2)
assert float(nll(.5,.5,.5,.5)) == likelyhood_answer
%%timeit
float(nll(*np.random.rand(4)))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
exit()
%run common.py
import torch
device = torch.device('cuda:0')
%%time
d_dist = torch.tensor(dist, device=device)
def gaussian(x, μ, σ):
return 1/np.sqrt(2*np.pi*σ**2) * torch.exp(-(x-μ)**2/(2*σ**2))
def add(x, f_0, mean, sigma, sigma2):
return f_0 * gaussian(x, mean, sigma) + (1 - f_0) * gaussian(x, mean, sigma2)
def nll(f_0, mean, sigma, sigma2):
return -torch.sum(torch.log(add(d_dist, f_0, mean, sigma, sigma2)))
v = nll(.5,.5,.5,.5)
assert v.item() == likelyhood_answer
%%timeit
nll(*np.random.rand(4))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
exit()
%run common.py
import tensorflow as tf
def gaussian(x, μ, σ):
return 1/tf.sqrt(2*np.pi*σ**2) * tf.math.exp(-(x-μ)**2/(2*σ**2))
def add(x, f_0, mean, sigma, sigma2):
return f_0 * gaussian(x, mean, sigma) + (1 - f_0) * gaussian(x, mean, sigma2)
def make_nll(d_dist, f_0, mean, sigma, sigma2):
return -tf.reduce_sum(tf.math.log(add(d_dist, f_0, mean, sigma, sigma2)))
x = tf.constant(dist)
tf_f_0 = tf.placeholder(dtype=tf.float64)
tf_mean = tf.placeholder(dtype=tf.float64)
tf_sigma = tf.placeholder(dtype=tf.float64)
tf_sigma2 = tf.placeholder(dtype=tf.float64)
graph = make_nll(x, tf_f_0, tf_mean, tf_sigma, tf_sigma2)
sess = tf.Session()
def nll(f_0, mean, sigma, sigma2):
loss_value = sess.run(graph,
feed_dict={tf_f_0:f_0,
tf_mean:mean,
tf_sigma:sigma,
tf_sigma2:sigma2})
return loss_value
assert nll(.5,.5,.5,.5) == likelyhood_answer
Using random numbers here just to make sure we don't get some special caching or somthing from TensorFlow.
%%timeit
nll(*np.random.rand(4))
minuit = Minuit(nll,
errordef=1,
**default_params)
%%time
run_and_print(minuit)
sess.close()
When using tensorflow, be a good user and kill the kernel so GPU memory gets reclaimed.
exit()
Quick check of summation algorithms:
>>> import numpy as np
>>> import math
>>> np.random.seed(42)
>>> dist = np.hstack([
... np.random.normal(loc=1, scale=2., size=500_000),
... np.random.normal(loc=1, scale=.5, size=500_000)
... ])
# fsum vs. naive
>>> math.fsum(dist) - sum(dist)
5.820766091346741e-10
# fsum vs. np.sum
>>> math.fsum(dist) - np.sum(dist)
0.0
# Check with 32 bit, fsum vs. naive
>>> math.fsum(dist.astype(np.float32)) - sum(dist.astype(np.float32), np.float32(0))
-20.27040922746528
# Check with 32 bit, fsum vs. np.sum
>>> math.fsum(dist.astype(np.float32)) - np.sum(dist.astype(np.float32))
-0.1454092274652794
# Check with 32 bit fsum vs. 64 bit fsum
>>> math.fsum(dist) - math.fsum(dist.astype(np.float32))
-2.3222528398036957e-06